76 std::istringstream iss(newValue);
90 double dimensionedReactionRate = reactionRate * (1e-3 *
m3 / (
mole *
s));
118 while(iss.eof() ==
false)
255 std::istringstream iss(newValue);
290 && iss.eof() == false
309 iss >> rateconst_method;
310 if(rateconst_method ==
"Fix")
316 double dimensionedReactionRate = reactionRate * (1e-3 *
m3 / (
mole *
s));
339 else if(rateconst_method ==
"Arr")
349 else if(rateconst_method ==
"Pol")
352 std::vector<double>
P = {0, 0, 0, 0, 0};
367 else if(rateconst_method ==
"Scale")
372 double reactionRateCste;
373 iss >> reactionRateCste;
374 double dimensionedReactionRate = reactionRateCste * (1e-3 *
m3 / (
mole *
s));
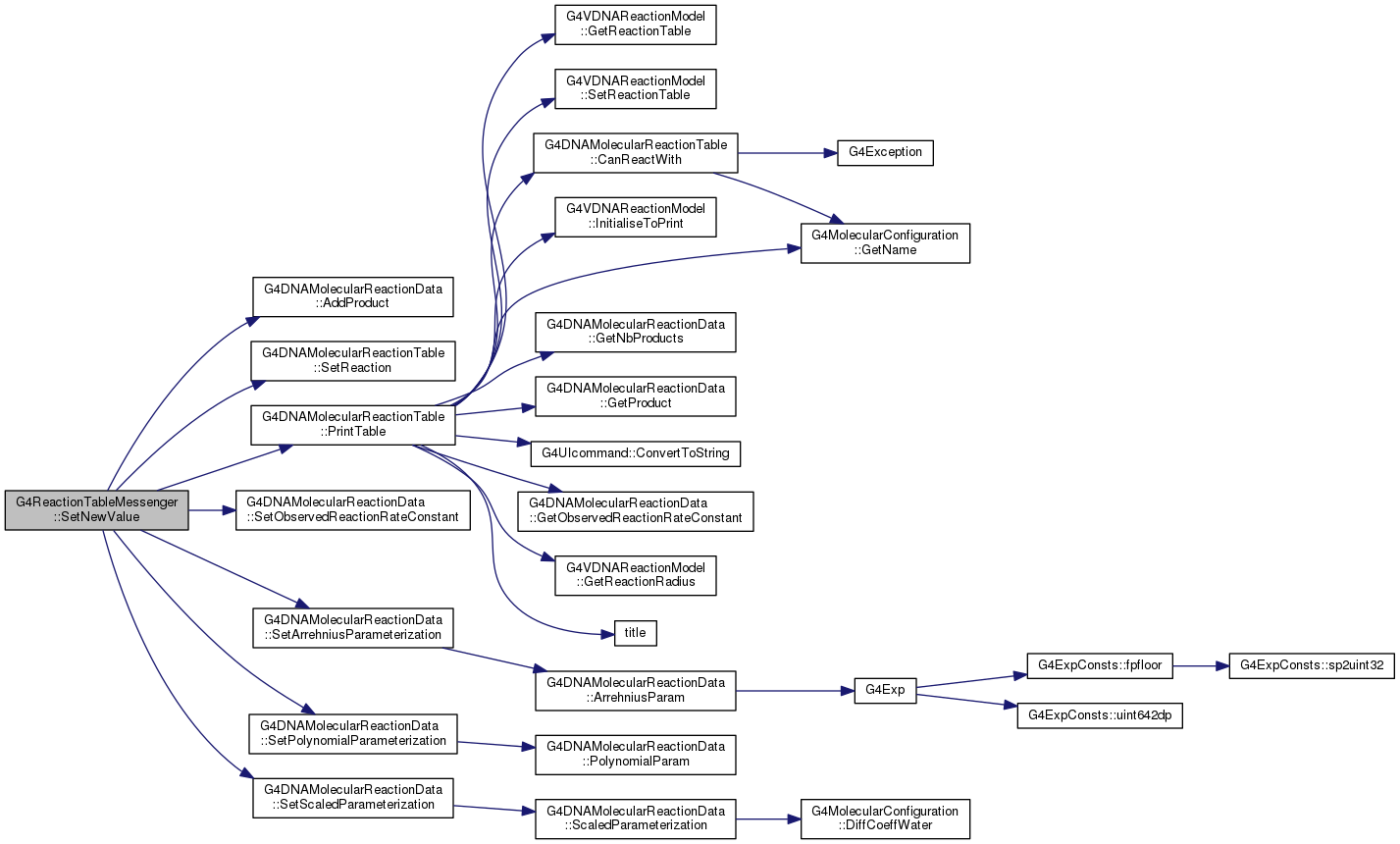
G4DNAMolecularReactionTable * fpTable
static constexpr double m3
void SetReaction(G4double observedReactionRate, G4MolecularConfiguration *reactive1, G4MolecularConfiguration *reactive2)
G4GLOB_DLL std::ostream G4cout
void SetArrehniusParameterization(double A0, double E_R)
void PrintTable(G4VDNAReactionModel *=0)
void AddProduct(G4MolecularConfiguration *molecule)
G4UIcmdWithAString * fpAddReaction
G4UIcmdWithoutParameter * fpPrintTable
void SetObservedReactionRateConstant(G4double rate)
G4UIcmdWithAString * fpNewDiffContReaction
void SetPolynomialParameterization(const std::vector< double > &P)
void SetScaledParameterization(double temperature_K, double rateCste)
static constexpr double mole
 Public Member Functions inherited from G4UImessenger
Public Member Functions inherited from G4UImessenger Protected Attributes inherited from G4UImessenger
Protected Attributes inherited from G4UImessenger Protected Member Functions inherited from G4UImessenger
Protected Member Functions inherited from G4UImessenger