|
Geant4
10.03.p03
|
|
Geant4
10.03.p03
|
#include <G4DNAMolecularReactionTable.hh>
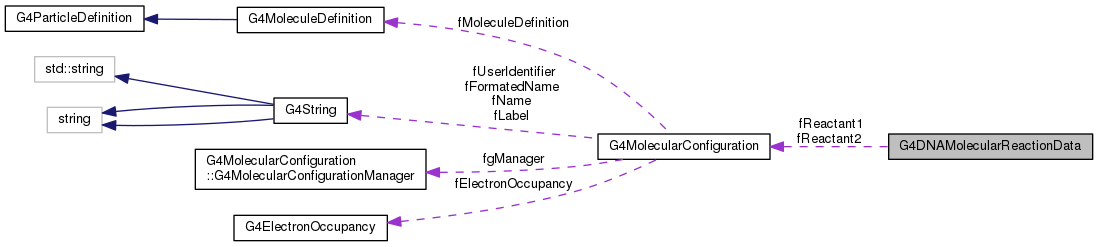
Public Types | |
| typedef std::function< double(double)> | RateParam |
Static Public Member Functions | |
| static double | PolynomialParam (double temp_K, std::vector< double > P) |
| static double | ArrehniusParam (double temp_K, std::vector< double > P) |
| static double | ScaledParameterization (double temp_K, double temp_init, double rateCste_init) |
Protected Member Functions | |
| G4DNAMolecularReactionData () | |
Protected Attributes | |
| G4MolecularConfiguration * | fReactant1 |
| G4MolecularConfiguration * | fReactant2 |
| G4double | fObservedReactionRate |
| G4double | fEffectiveReactionRadius |
| std::vector < G4MolecularConfiguration * > * | fProducts |
| RateParam | fRateParam |
| int | fReactionID |
G4DNAMolecularReactionData contains the information relative to a given reaction (eg : °OH + °OH -> H2O2)
Definition at line 67 of file G4DNAMolecularReactionTable.hh.
| typedef std::function<double(double)> G4DNAMolecularReactionData::RateParam |
Definition at line 165 of file G4DNAMolecularReactionTable.hh.
| G4DNAMolecularReactionData::G4DNAMolecularReactionData | ( | G4double | reactionRate, |
| G4MolecularConfiguration * | reactive1, | ||
| G4MolecularConfiguration * | reactive2 | ||
| ) |
Definition at line 70 of file G4DNAMolecularReactionTable.cc.
| G4DNAMolecularReactionData::G4DNAMolecularReactionData | ( | G4double | reactionRate, |
| const G4String & | reactive1, | ||
| const G4String & | reactive2 | ||
| ) |
Definition at line 100 of file G4DNAMolecularReactionTable.cc.
| G4DNAMolecularReactionData::~G4DNAMolecularReactionData | ( | ) |
Definition at line 127 of file G4DNAMolecularReactionTable.cc.
|
protected |
Definition at line 57 of file G4DNAMolecularReactionTable.cc.
| void G4DNAMolecularReactionData::AddProduct | ( | G4MolecularConfiguration * | molecule | ) |
Definition at line 155 of file G4DNAMolecularReactionTable.cc.
Definition at line 176 of file G4DNAMolecularReactionTable.cc.

|
static |
Definition at line 193 of file G4DNAMolecularReactionTable.cc.

|
inline |
Definition at line 112 of file G4DNAMolecularReactionTable.hh.
|
inline |
Definition at line 137 of file G4DNAMolecularReactionTable.hh.
|
inline |
Definition at line 107 of file G4DNAMolecularReactionTable.hh.
|
inline |
Definition at line 143 of file G4DNAMolecularReactionTable.hh.
|
inline |
Definition at line 149 of file G4DNAMolecularReactionTable.hh.
|
inline |
Definition at line 93 of file G4DNAMolecularReactionTable.hh.
|
inline |
Definition at line 97 of file G4DNAMolecularReactionTable.hh.
|
inline |
Definition at line 88 of file G4DNAMolecularReactionTable.hh.
|
inline |
Definition at line 83 of file G4DNAMolecularReactionTable.hh.
|
static |
Definition at line 183 of file G4DNAMolecularReactionTable.cc.

|
inline |
Definition at line 154 of file G4DNAMolecularReactionTable.hh.
|
static |
Definition at line 199 of file G4DNAMolecularReactionTable.cc.

| void G4DNAMolecularReactionData::ScaleForNewTemperature | ( | double | temp_K | ) |
Definition at line 726 of file G4DNAMolecularReactionTable.cc.
| void G4DNAMolecularReactionData::SetArrehniusParameterization | ( | double | A0, |
| double | E_R | ||
| ) |
Definition at line 689 of file G4DNAMolecularReactionTable.cc.

Definition at line 117 of file G4DNAMolecularReactionTable.hh.
Definition at line 102 of file G4DNAMolecularReactionTable.hh.
| void G4DNAMolecularReactionData::SetPolynomialParameterization | ( | const std::vector< double > & | P | ) |
Definition at line 682 of file G4DNAMolecularReactionTable.cc.
| void G4DNAMolecularReactionData::SetReactant1 | ( | G4MolecularConfiguration * | reactive | ) |
Definition at line 137 of file G4DNAMolecularReactionTable.cc.
Definition at line 161 of file G4DNAMolecularReactionTable.cc.
| void G4DNAMolecularReactionData::SetReactant2 | ( | G4MolecularConfiguration * | reactive | ) |
Definition at line 143 of file G4DNAMolecularReactionTable.cc.
Definition at line 165 of file G4DNAMolecularReactionTable.cc.
| void G4DNAMolecularReactionData::SetReactants | ( | G4MolecularConfiguration * | reactive1, |
| G4MolecularConfiguration * | reactive2 | ||
| ) |
Definition at line 148 of file G4DNAMolecularReactionTable.cc.
| void G4DNAMolecularReactionData::SetReactants | ( | const G4String & | reactive1, |
| const G4String & | reactive2 | ||
| ) |
Definition at line 169 of file G4DNAMolecularReactionTable.cc.
Definition at line 84 of file G4DNAMolecularReactionTable.hh.
| void G4DNAMolecularReactionData::SetScaledParameterization | ( | double | temperature_K, |
| double | rateCste | ||
| ) |
Definition at line 702 of file G4DNAMolecularReactionTable.cc.

|
protected |
Definition at line 186 of file G4DNAMolecularReactionTable.hh.
|
protected |
Definition at line 185 of file G4DNAMolecularReactionTable.hh.
|
protected |
Definition at line 188 of file G4DNAMolecularReactionTable.hh.
|
protected |
Definition at line 190 of file G4DNAMolecularReactionTable.hh.
|
protected |
Definition at line 183 of file G4DNAMolecularReactionTable.hh.
|
protected |
Definition at line 184 of file G4DNAMolecularReactionTable.hh.
|
protected |
Definition at line 191 of file G4DNAMolecularReactionTable.hh.