#include <G4EmDNAChemistry.hh>
Definition at line 35 of file G4EmDNAChemistry.hh.
| G4EmDNAChemistry::G4EmDNAChemistry |
( |
| ) |
|
|
explicit |
Definition at line 92 of file G4EmDNAChemistry.cc.
void SetChemistryList(G4VUserChemistryList *)
G4VUserChemistryList(bool flag=true)
static G4DNAChemistryManager * Instance()
| G4EmDNAChemistry::~G4EmDNAChemistry |
( |
| ) |
|
|
virtual |
| void G4EmDNAChemistry::ConstructDissociationChannels |
( |
| ) |
|
|
virtual |
Reimplemented from G4VUserChemistryList.
Definition at line 142 of file G4EmDNAChemistry.cc.
186 G4DNAWaterDissociationDisplacer::A1B1_DissociationDecay);
201 "B^1A_1_AutoIonisation_Channel");
213 G4DNAWaterDissociationDisplacer::B1A1_DissociationDecay);
234 "Excitation3rdLayer_AutoIonisation_Channel");
236 "Excitation3rdLayer_Relaxation_Channel");
263 "Excitation2ndLayer_AutoIonisation_Channel");
265 "Excitation2ndLayer_Relaxation_Channel");
290 "Excitation1stLayer_AutoIonisation_Channel");
292 "Excitation1stLayer_Relaxation_Channel");
326 G4DNAWaterDissociationDisplacer::Ionisation_DissociationDecay);
370 DissociativeAttachment);
G4double ExcitationEnergy(G4int level)
static G4H2O * Definition()
void SetProbability(G4double)
void AddDecayChannel(const G4MolecularConfiguration *molConf, const G4MolecularDissociationChannel *channel)
void AddProduct(const G4Molecule *, G4double=0)
G4int AddElectron(G4int orbit, G4int number=1)
static G4MoleculeTable * Instance()
const G4ElectronOccupancy * GetGroundStateElectronOccupancy() const
G4MolecularConfiguration * NewConfigurationWithElectronOccupancy(const G4String &excitedStateLabel, const G4ElectronOccupancy &, double decayTime=0.)
void SetDisplacementType(DisplacementType)
G4int RemoveElectron(G4int orbit, G4int number=1)
G4MolecularConfiguration * GetConfiguration(const G4String &, bool mustExist=true)
| void G4EmDNAChemistry::ConstructMolecule |
( |
| ) |
|
|
virtual |
Reimplemented from G4VUserChemistryList.
Definition at line 106 of file G4EmDNAChemistry.cc.
125 CreateConfiguration(
"OHm",
static G4Electron_aq * Definition()
static G4H2O * Definition()
static G4Electron * Definition()
static constexpr double g
static constexpr double Avogadro
static G4H3O * Definition()
static G4MoleculeTable * Instance()
static constexpr double c_squared
G4MolecularConfiguration * CreateConfiguration(const G4String &userIdentifier, const G4MoleculeDefinition *molDef, const G4String &configurationLabel, const G4ElectronOccupancy &eOcc)
static G4H2O2 * Definition()
static G4OH * Definition()
static G4H2 * Definition()
static constexpr double m2
static G4Hydrogen * Definition()
| virtual void G4EmDNAChemistry::ConstructParticle |
( |
| ) |
|
|
inlinevirtual |
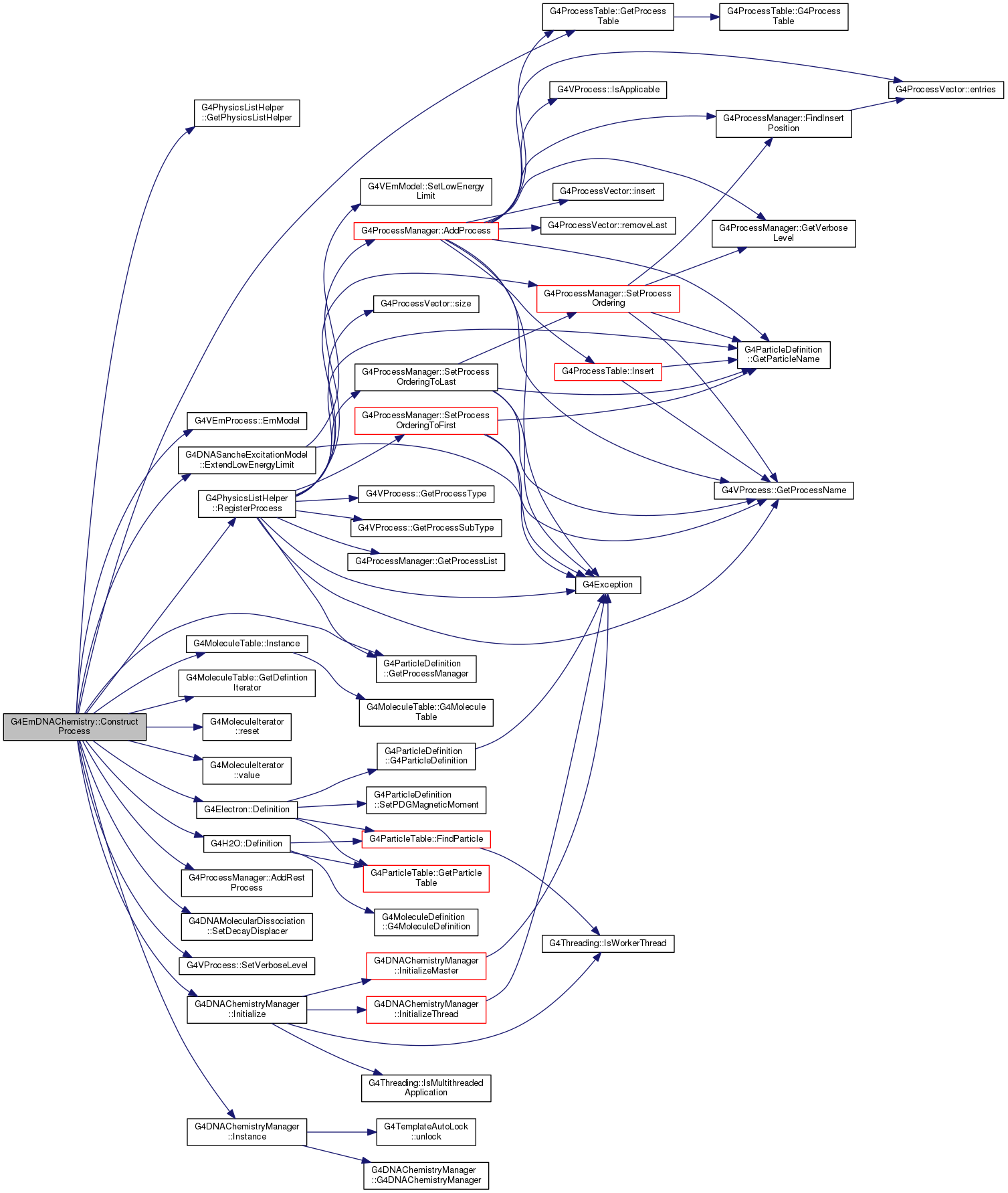
| void G4EmDNAChemistry::ConstructProcess |
( |
| ) |
|
|
virtual |
Reimplemented from G4VUserChemistryList.
Definition at line 459 of file G4EmDNAChemistry.cc.
470 FindProcess(
"e-_G4DNAVibExcitation",
"e-");
478 if(sancheExcitationMod)
489 FindProcess(
"e-_G4DNAElectronSolvation",
"e-");
static G4H2O * Definition()
G4VEmModel * EmModel(G4int index=1) const
void ExtendLowEnergyLimit(G4double)
static G4Electron * Definition()
G4bool RegisterProcess(G4VProcess *process, G4ParticleDefinition *particle)
static G4MoleculeTable * Instance()
static constexpr double eV
static G4DNAChemistryManager * Instance()
G4MoleculeDefinitionIterator GetDefintionIterator()
G4ProcessManager * GetProcessManager() const
static G4PhysicsListHelper * GetPhysicsListHelper()
G4int AddRestProcess(G4VProcess *aProcess, G4int ord=ordDefault)
void SetDecayDisplacer(const G4ParticleDefinition *, G4VMolecularDecayDisplacer *)
const XML_Char XML_Content * model
static G4ProcessTable * GetProcessTable()
void SetVerboseLevel(G4int value)
Implements G4VUserChemistryList.
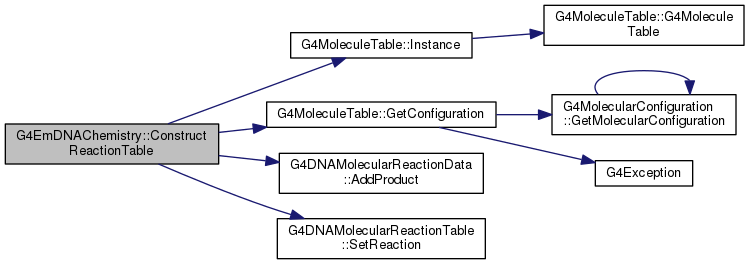
Definition at line 382 of file G4EmDNAChemistry.cc.
409 theReactionTable->SetReaction(reactionData);
413 2.95e10 * (1e-3 *
m3 / (
mole *
s)), e_aq, OH);
415 theReactionTable->SetReaction(reactionData);
419 2.65e10 * (1e-3 *
m3 / (
mole *
s)), e_aq, H);
422 theReactionTable->SetReaction(reactionData);
426 2.11e10 * (1e-3 *
m3 / (
mole *
s)), e_aq, H3Op);
428 theReactionTable->SetReaction(reactionData);
432 1.41e10 * (1e-3 *
m3 / (
mole *
s)), e_aq, H2O2);
435 theReactionTable->SetReaction(reactionData);
439 0.44e10 * (1e-3 *
m3 / (
mole *
s)), OH, OH);
441 theReactionTable->SetReaction(reactionData);
444 theReactionTable->SetReaction(1.44e10 * (1e-3 *
m3 / (
mole *
s)), OH, H);
448 1.20e10 * (1e-3 *
m3 / (
mole * s)), H, H);
450 theReactionTable->SetReaction(reactionData);
453 theReactionTable->SetReaction(1.43e11 * (1e-3 *
m3 / (
mole * s)), H3Op, OHm);
static constexpr double m3
static G4MoleculeTable * Instance()
void AddProduct(G4MolecularConfiguration *molecule)
static constexpr double mole
G4MolecularConfiguration * GetConfiguration(const G4String &, bool mustExist=true)
The reaction model defines how to compute the reaction range between molecules
The StepByStep model tells the step manager how to behave before and after each step, how to compute the time steps.
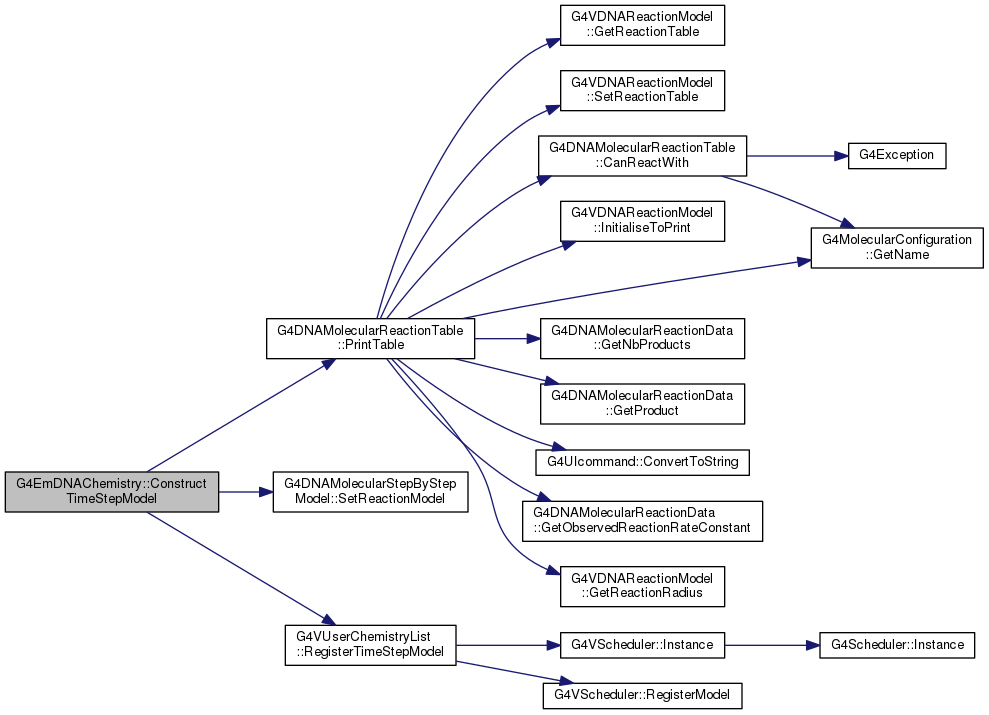
Implements G4VUserChemistryList.
Definition at line 542 of file G4EmDNAChemistry.cc.
556 reactionTable->
PrintTable(reactionRadiusComputer);
void PrintTable(G4VDNAReactionModel *=0)
void SetReactionModel(G4VDNAReactionModel *)
void RegisterTimeStepModel(G4VITStepModel *timeStepModel, double startingTime=0)
The documentation for this class was generated from the following files:
 Static Public Member Functions inherited from G4VPhysicsConstructor
Static Public Member Functions inherited from G4VPhysicsConstructor Protected Member Functions inherited from G4VUserChemistryList
Protected Member Functions inherited from G4VUserChemistryList Protected Member Functions inherited from G4VPhysicsConstructor
Protected Member Functions inherited from G4VPhysicsConstructor Protected Attributes inherited from G4VUserChemistryList
Protected Attributes inherited from G4VUserChemistryList Protected Attributes inherited from G4VPhysicsConstructor
Protected Attributes inherited from G4VPhysicsConstructor Static Protected Attributes inherited from G4VPhysicsConstructor
Static Protected Attributes inherited from G4VPhysicsConstructor