|
Geant4
10.03.p03
|
|
Geant4
10.03.p03
|
#include <ScoreSpecies.hh>
Classes | |
| struct | SpeciesInfo |
Protected Member Functions | |
| virtual G4bool | ProcessHits (G4Step *, G4TouchableHistory *) |
 Protected Member Functions inherited from G4VPrimitiveScorer Protected Member Functions inherited from G4VPrimitiveScorer | |
| virtual G4int | GetIndex (G4Step *) |
| void | CheckAndSetUnit (const G4String &unit, const G4String &category) |
Additional Inherited Members | |
 Protected Attributes inherited from G4VPrimitiveScorer Protected Attributes inherited from G4VPrimitiveScorer | |
| G4String | primitiveName |
| G4MultiFunctionalDetector * | detector |
| G4VSDFilter * | filter |
| G4int | verboseLevel |
| G4int | indexDepth |
| G4String | unitName |
| G4double | unitValue |
| G4int | fNi |
| G4int | fNj |
| G4int | fNk |
This is a primitive scorer class for molecular species. The number of species is recorded for all times (predetermined or user chosen). It also scores the energy deposition in order to compute the radiochemical yields.
Definition at line 56 of file ScoreSpecies.hh.
Definition at line 62 of file ScoreSpecies.cc.

|
virtual |
Definition at line 84 of file ScoreSpecies.cc.
|
virtual |
Method used in multithreading mode in order to merge the results
Definition at line 198 of file ScoreSpecies.cc.

|
inline |
Add a time at which the number of species should be recorded. Default times are set up to 1 microsecond.
Definition at line 65 of file ScoreSpecies.hh.

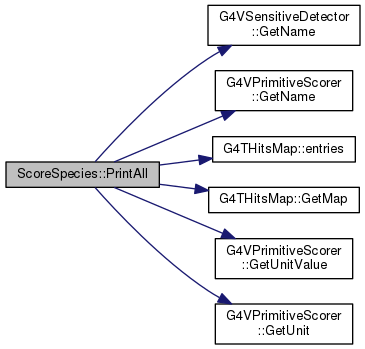
| void ScoreSpecies::ASCII | ( | ) |
Reimplemented from G4VPrimitiveScorer.
Definition at line 285 of file ScoreSpecies.cc.


|
inline |
Remove all times to record, must be reset by user.
Definition at line 71 of file ScoreSpecies.hh.
|
virtual |
Reimplemented from G4VPrimitiveScorer.
Definition at line 120 of file ScoreSpecies.cc.
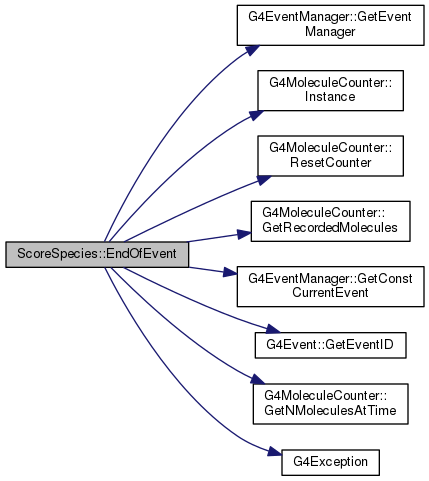
|
inline |
Get number of recorded events
Definition at line 77 of file ScoreSpecies.hh.
|
inline |
Definition at line 181 of file ScoreSpecies.hh.
|
virtual |
Reimplemented from G4VPrimitiveScorer.
Definition at line 105 of file ScoreSpecies.cc.
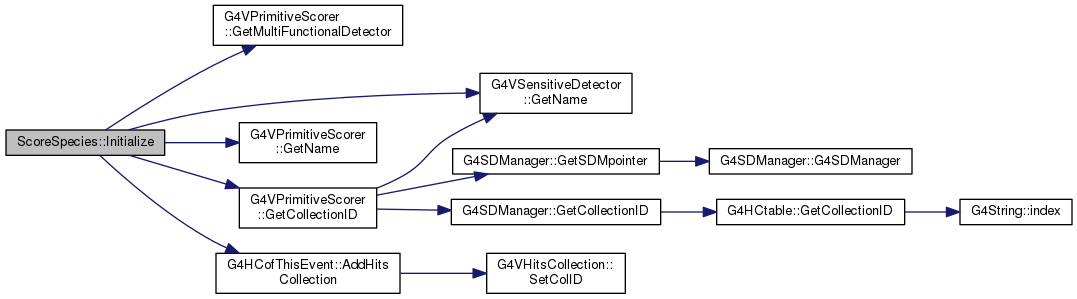
|
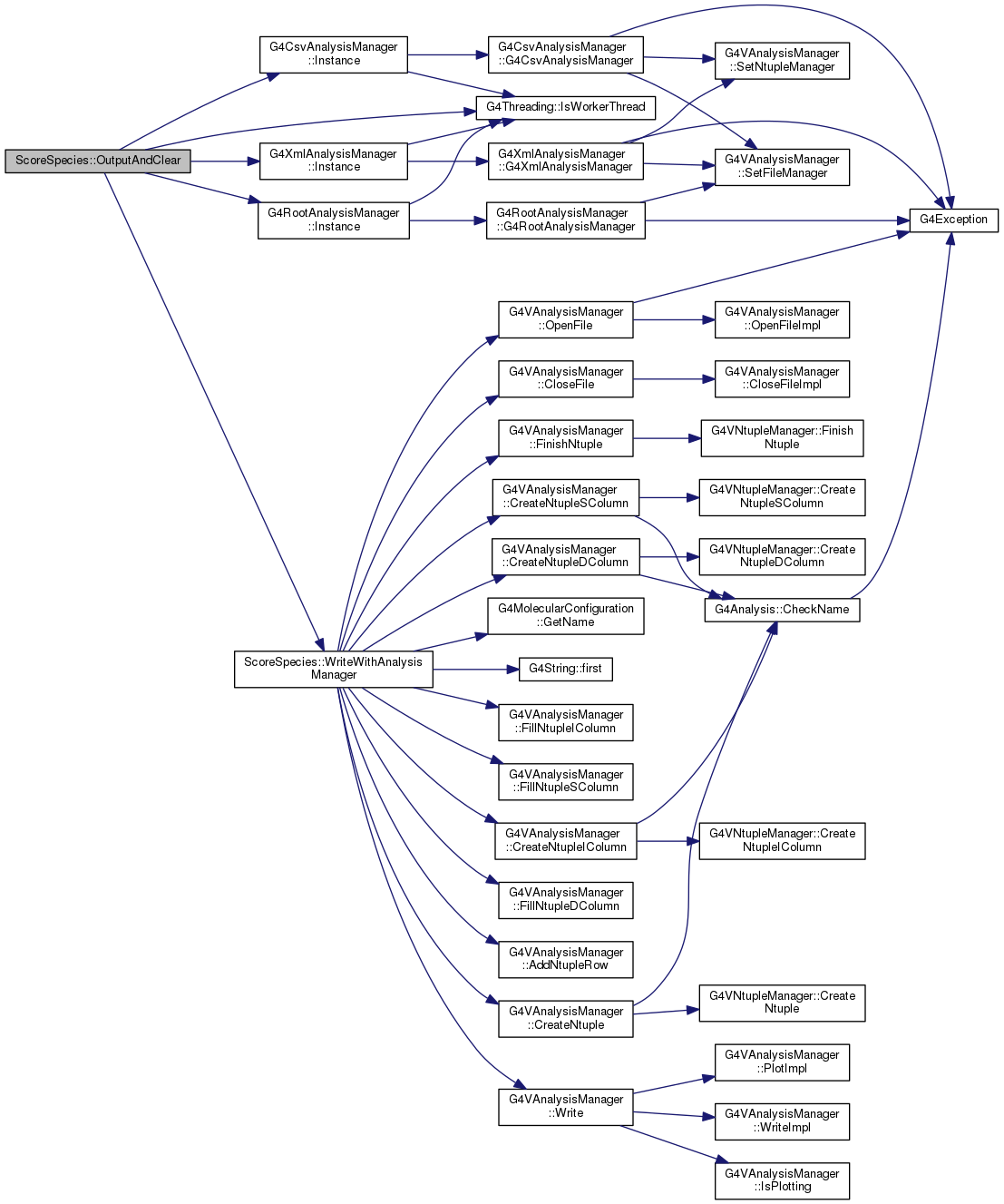
virtual |
Definition at line 344 of file ScoreSpecies.cc.
Reimplemented from G4VPrimitiveScorer.
Definition at line 300 of file ScoreSpecies.cc.
|
protectedvirtual |
Implements G4VPrimitiveScorer.
Definition at line 89 of file ScoreSpecies.cc.
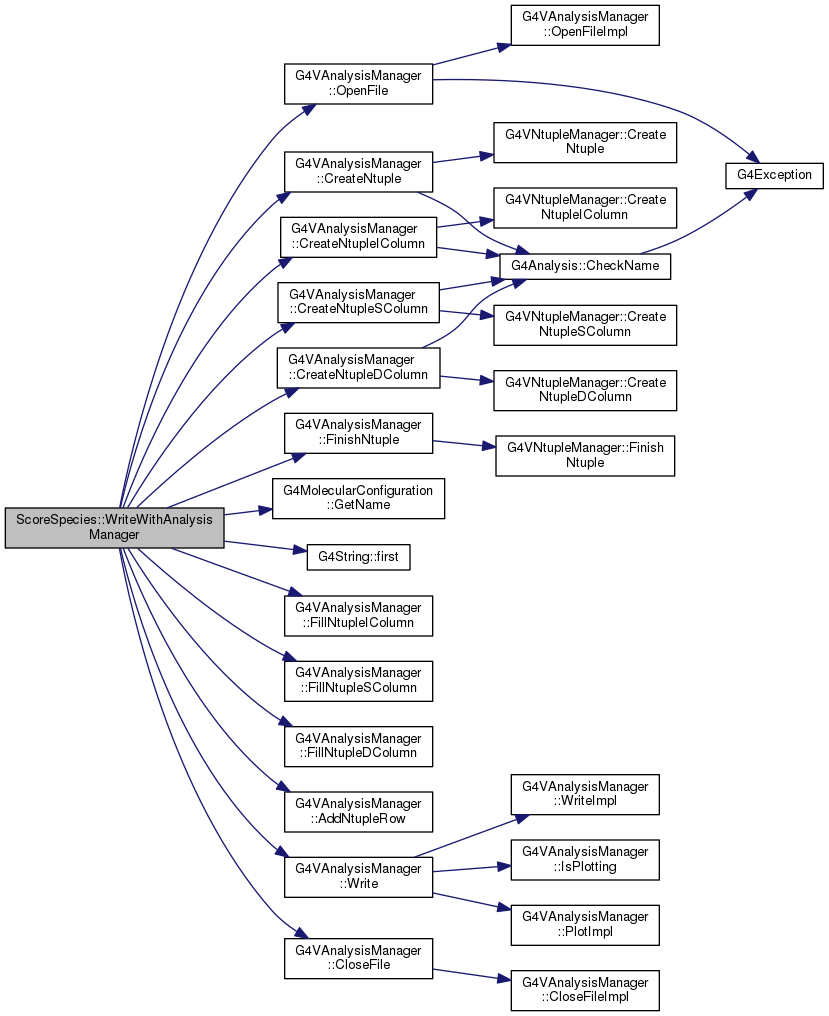
| void ScoreSpecies::WriteWithAnalysisManager | ( | G4VAnalysisManager * | analysisManager | ) |
Write results to whatever chosen file format
Definition at line 379 of file ScoreSpecies.cc.