|
Geant4
10.03.p03
|
|
Geant4
10.03.p03
|
#include <G4DNAChemistryManager.hh>
Static Public Member Functions | |
| static G4DNAChemistryManager * | Instance () |
| static G4DNAChemistryManager * | GetInstanceIfExists () |
| static void | DeleteInstance () |
| static G4bool | IsActivated () |
| static void | Activated (G4bool flag=true) |
Protected Member Functions | |
| virtual | ~G4DNAChemistryManager () |
| G4DNAWaterExcitationStructure * | GetExcitationLevel () |
| G4DNAWaterIonisationStructure * | GetIonisationLevel () |
| void | InitializeFile () |
| void | InitializeMaster () |
| void | InitializeThread () |
| G4DNAChemistryManager () | |
 Protected Member Functions inherited from G4UImessenger Protected Member Functions inherited from G4UImessenger | |
| G4String | ItoS (G4int i) |
| G4String | DtoS (G4double a) |
| G4String | BtoS (G4bool b) |
| G4int | StoI (G4String s) |
| G4double | StoD (G4String s) |
| G4bool | StoB (G4String s) |
| void | AddUIcommand (G4UIcommand *newCommand) |
| void | CreateDirectory (const G4String &path, const G4String &dsc, G4bool commandsToBeBroadcasted=true) |
| template<typename T > | |
| T * | CreateCommand (const G4String &cname, const G4String &dsc) |
Additional Inherited Members | |
 Protected Attributes inherited from G4UImessenger Protected Attributes inherited from G4UImessenger | |
| G4UIdirectory * | baseDir |
| G4String | baseDirName |
| G4bool | commandsShouldBeInMaster |
WARNING: THIS CLASS IS A PROTOTYPE G4DNAChemistryManager is called from the physics models. It creates the water molecules and the solvated electrons and and send them to G4ITStepManager to be treated in the chemistry stage. For this, the fActiveChemistry flag needs to be on. It is also possible to give already molecule's pointers already built. G4DNAChemistryManager will then be in charge of creating the track and loading it to the IT system. The user can also ask to create a file containing a information about the creation of water molecules and solvated electrons.
Definition at line 88 of file G4DNAChemistryManager.hh.
|
protectedvirtual |
|
protected |
Definition at line 70 of file G4DNAChemistryManager.cc.
Definition at line 500 of file G4DNAChemistryManager.cc.

| void G4DNAChemistryManager::AddEmptyLineInOuputFile | ( | ) |
Definition at line 530 of file G4DNAChemistryManager.cc.
| void G4DNAChemistryManager::Clear | ( | ) |
Definition at line 149 of file G4DNAChemistryManager.cc.

| void G4DNAChemistryManager::CloseFile | ( | ) |
Close the file specified with WriteInto
Definition at line 538 of file G4DNAChemistryManager.cc.
| void G4DNAChemistryManager::CreateSolvatedElectron | ( | const G4Track * | theIncomingTrack, |
| G4ThreeVector * | finalPosition = 0 |
||
| ) |
On the same idea as the previous method but for solvated electron. This method should be used by the physics model of the ElectronSolvatation process.
Definition at line 638 of file G4DNAChemistryManager.cc.
| void G4DNAChemistryManager::CreateWaterMolecule | ( | ElectronicModification | modification, |
| G4int | electronicLevel, | ||
| const G4Track * | theIncomingTrack | ||
| ) |
Method used by DNA physics model to create a water molecule. The ElectronicModification is a flag telling wheter the molecule is ionized or excited, the electronic level is calculated by the model and the IncomingTrack is the track responsible for the creation of this molecule, for instance an electron.
Definition at line 572 of file G4DNAChemistryManager.cc.
|
static |
You should rather use DeleteInstance than the destructor of this class
Definition at line 209 of file G4DNAChemistryManager.cc.
|
inline |
Definition at line 269 of file G4DNAChemistryManager.hh.
|
inline |
Definition at line 274 of file G4DNAChemistryManager.hh.
|
inline |
Definition at line 258 of file G4DNAChemistryManager.hh.
|
inline |
Definition at line 280 of file G4DNAChemistryManager.hh.
|
virtual |
Reimplemented from G4UImessenger.
Definition at line 283 of file G4DNAChemistryManager.cc.
|
protected |
Definition at line 553 of file G4DNAChemistryManager.cc.
|
static |
Definition at line 132 of file G4DNAChemistryManager.cc.
|
protected |
Definition at line 563 of file G4DNAChemistryManager.cc.
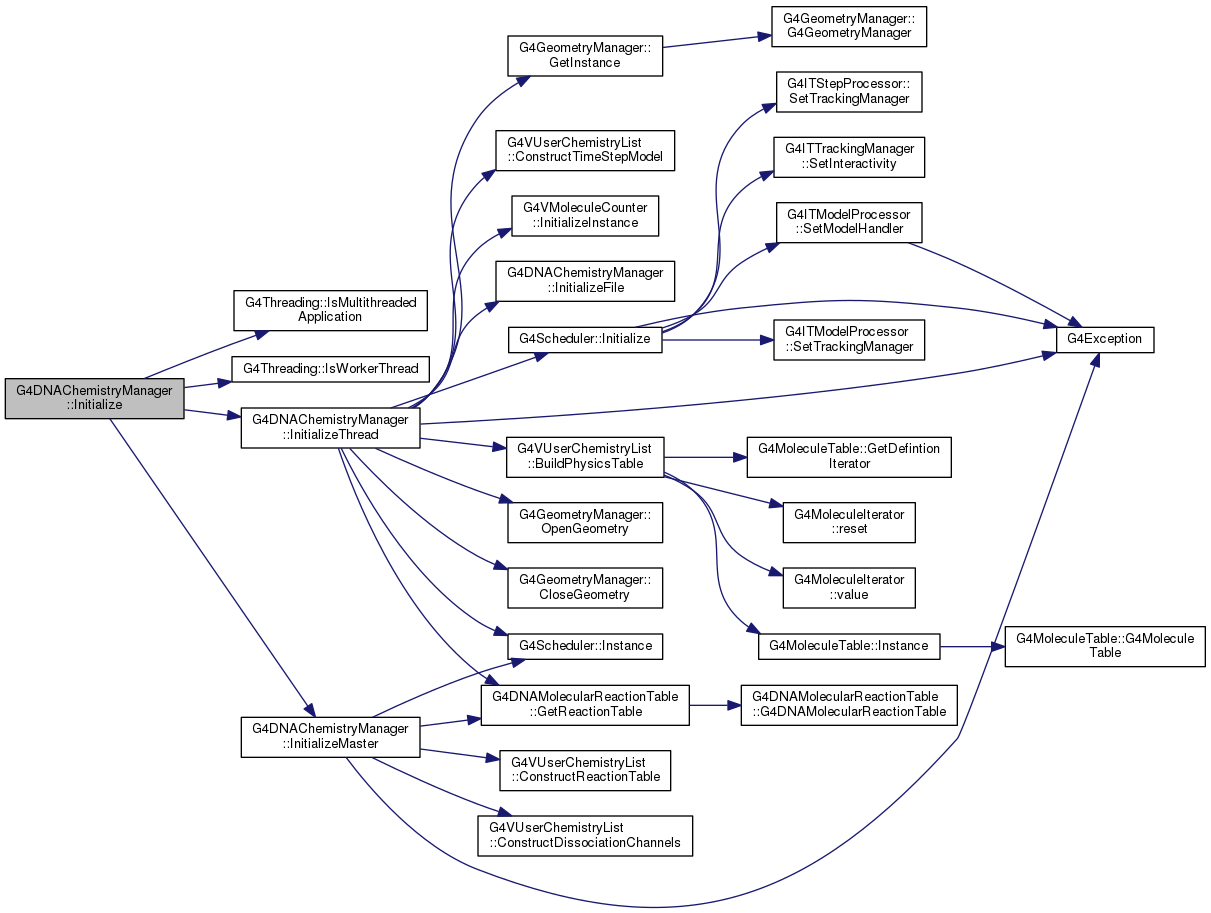
| void G4DNAChemistryManager::Initialize | ( | ) |
Definition at line 331 of file G4DNAChemistryManager.cc.
|
protected |
Definition at line 466 of file G4DNAChemistryManager.cc.

|
protected |
Definition at line 367 of file G4DNAChemistryManager.cc.
|
protected |
Definition at line 407 of file G4DNAChemistryManager.cc.
|
static |
Definition at line 117 of file G4DNAChemistryManager.cc.
|
static |
Definition at line 495 of file G4DNAChemistryManager.cc.
| G4bool G4DNAChemistryManager::IsChemistryActivated | ( | ) |
Definition at line 505 of file G4DNAChemistryManager.cc.
|
inline |
Definition at line 206 of file G4DNAChemistryManager.hh.
|
virtual |
Implements G4VStateDependent.
Definition at line 229 of file G4DNAChemistryManager.cc.
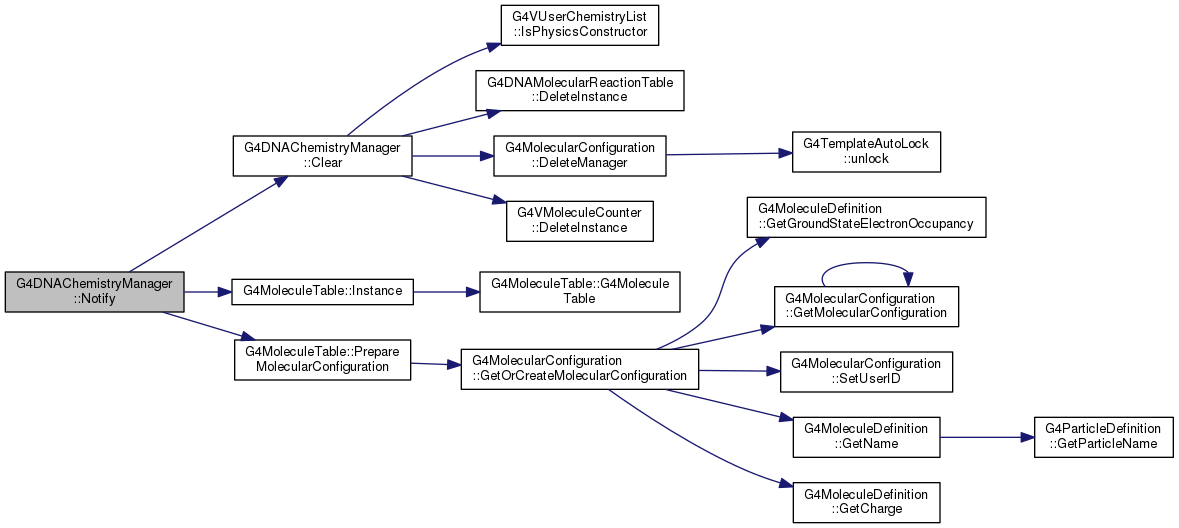
| void G4DNAChemistryManager::PushMolecule | ( | G4Molecule *& | molecule, |
| double | time, | ||
| const G4ThreeVector & | position, | ||
| int | parentID | ||
| ) |
WARNING : In case chemistry is not activated, PushMolecule will take care of deleting the transfered molecule. Before calling this method, it is also possible to check if the chemistry is activated through IsChemistryActived(). This method will create the track corresponding to the transfered molecule and will be in charge of loading the new track to the system.
Definition at line 685 of file G4DNAChemistryManager.cc.
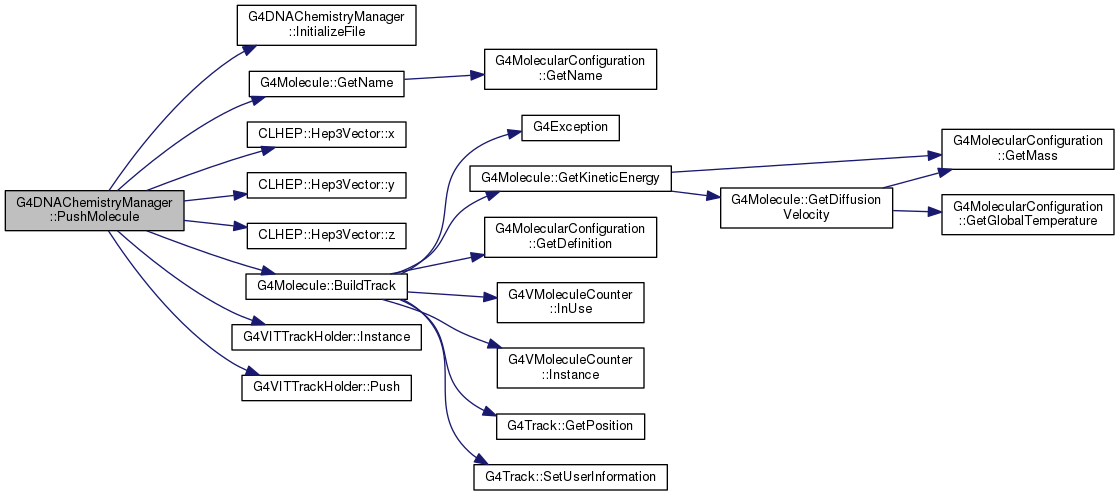
| void G4DNAChemistryManager::PushMoleculeAtParentTimeAndPlace | ( | G4Molecule *& | molecule, |
| const G4Track * | theIncomingTrack | ||
| ) |
WARNING : In case chemistry is not activated, PushMoleculeAtParentTimeAndPlace will take care of deleting the transfered molecule. Before calling this method, it is also possible to check if the chemistry is activated through IsChemistryActived(). This method will create the track corresponding to the transfered molecule and will be in charge of loading the new track to the system.
Definition at line 717 of file G4DNAChemistryManager.cc.
Definition at line 211 of file G4DNAChemistryManager.hh.
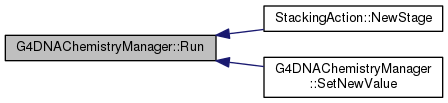
| void G4DNAChemistryManager::Run | ( | ) |
Definition at line 293 of file G4DNAChemistryManager.cc.
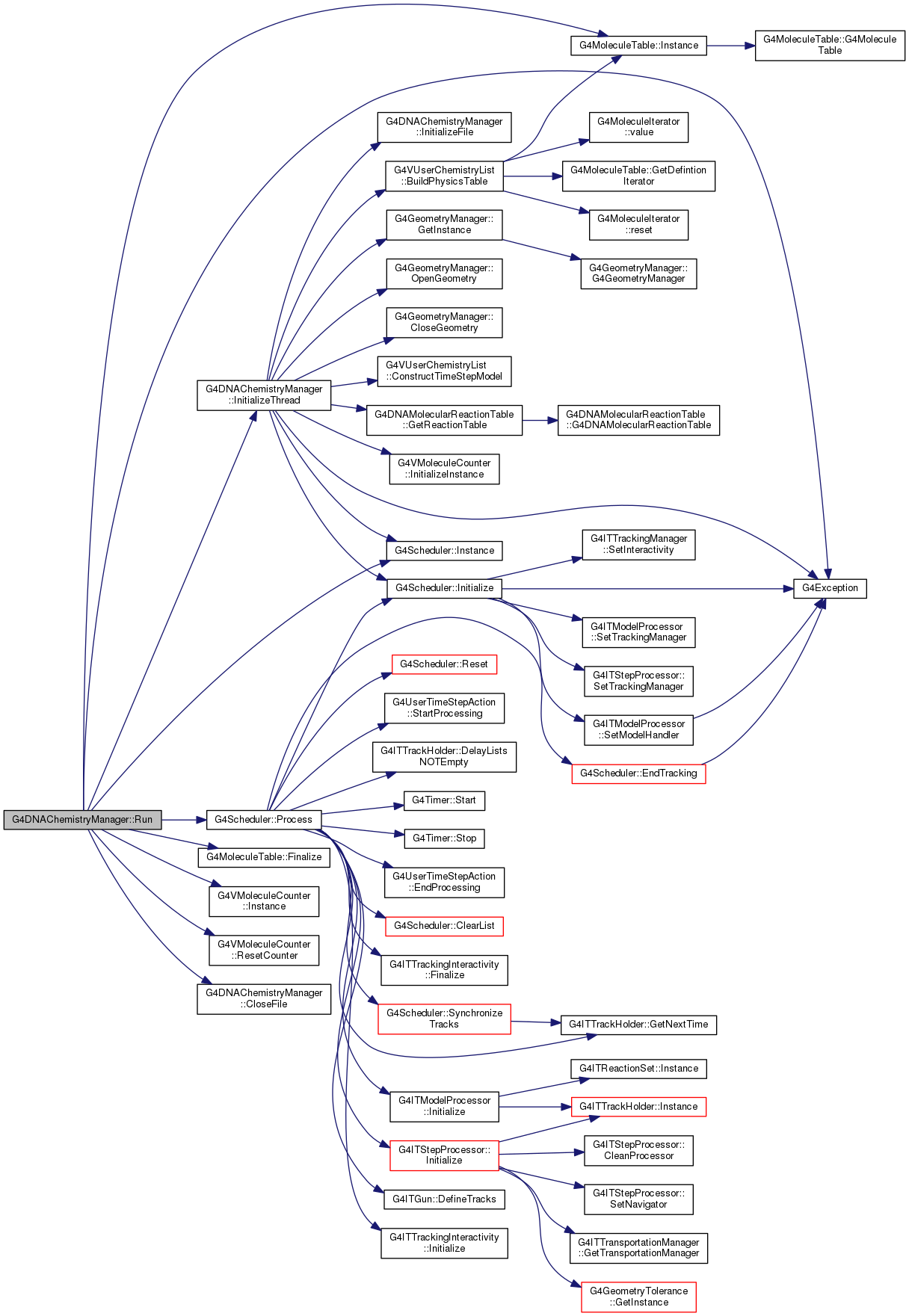
|
inline |
Definition at line 203 of file G4DNAChemistryManager.hh.
Definition at line 510 of file G4DNAChemistryManager.cc.
|
inline |
Definition at line 263 of file G4DNAChemistryManager.hh.
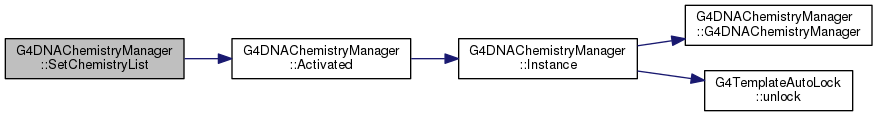

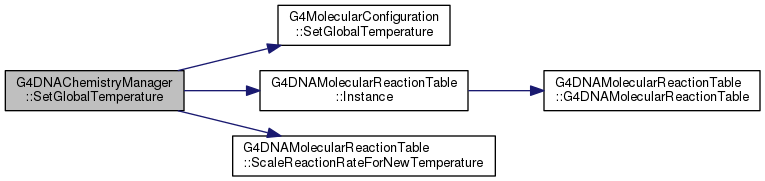
| void G4DNAChemistryManager::SetGlobalTemperature | ( | double | temp_K | ) |
Definition at line 752 of file G4DNAChemistryManager.cc.

|
virtual |
Reimplemented from G4UImessenger.
Definition at line 253 of file G4DNAChemistryManager.cc.
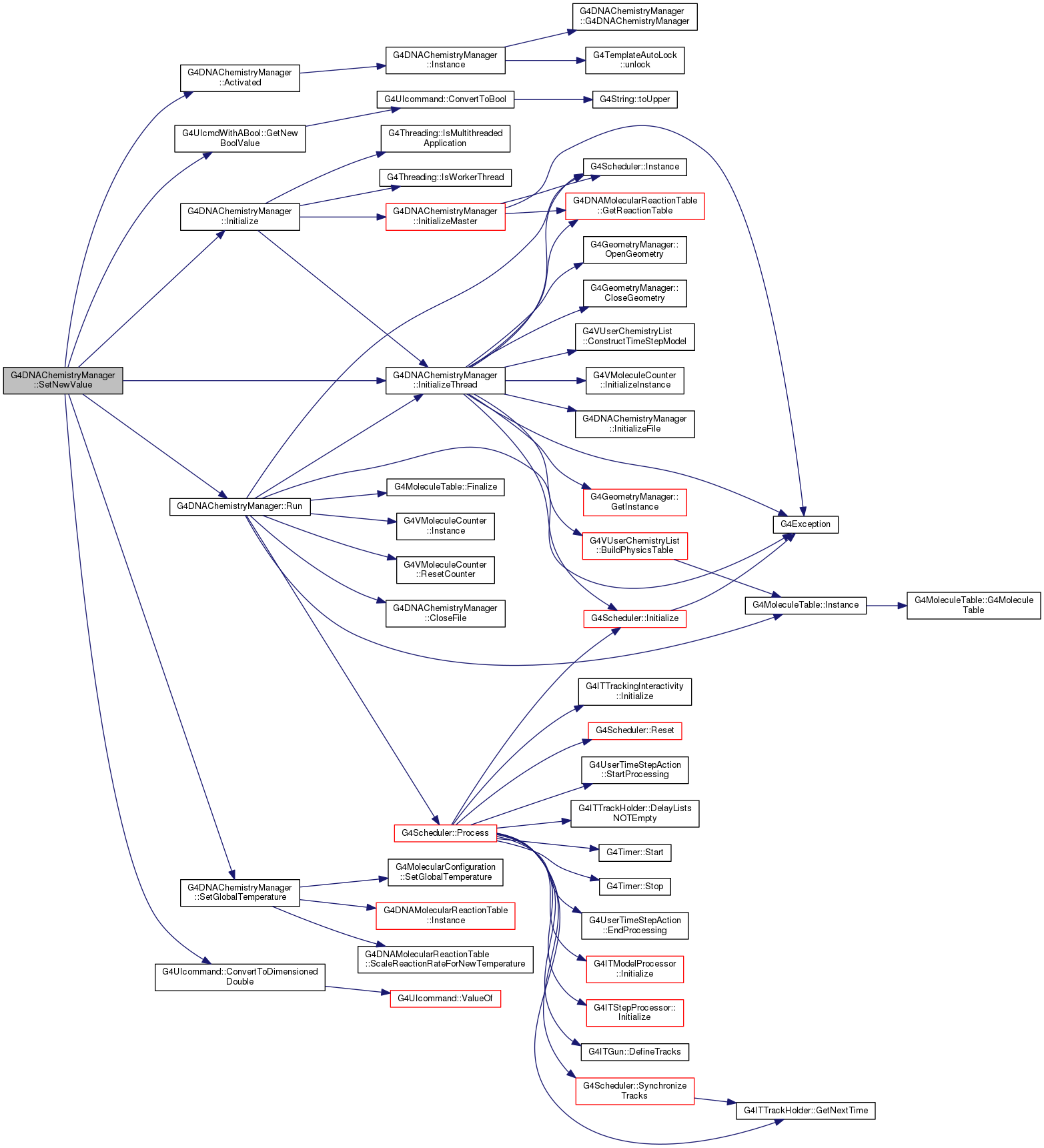
Definition at line 198 of file G4DNAChemistryManager.hh.
|
inline |
Definition at line 286 of file G4DNAChemistryManager.hh.
| void G4DNAChemistryManager::WriteInto | ( | const G4String & | , |
| std::ios_base::openmode | mode = std::ios_base::out |
||
| ) |
Tells the chemMan to write into a file the position and electronic state of the water molecule and the position thermalized or not of the solvated electron
Definition at line 515 of file G4DNAChemistryManager.cc.